MolT5: Translation between Molecules and Natural Language
Carl Edwards, Tuan Lai, Kevin Ros, Garrett Honke, Kyunghyun Cho, Heng Ji
Contact: hengji@illinois.edu, cne2@illinois.edu
Please email Carl Edwards if you experience any technical issues using our software or need further information.
About
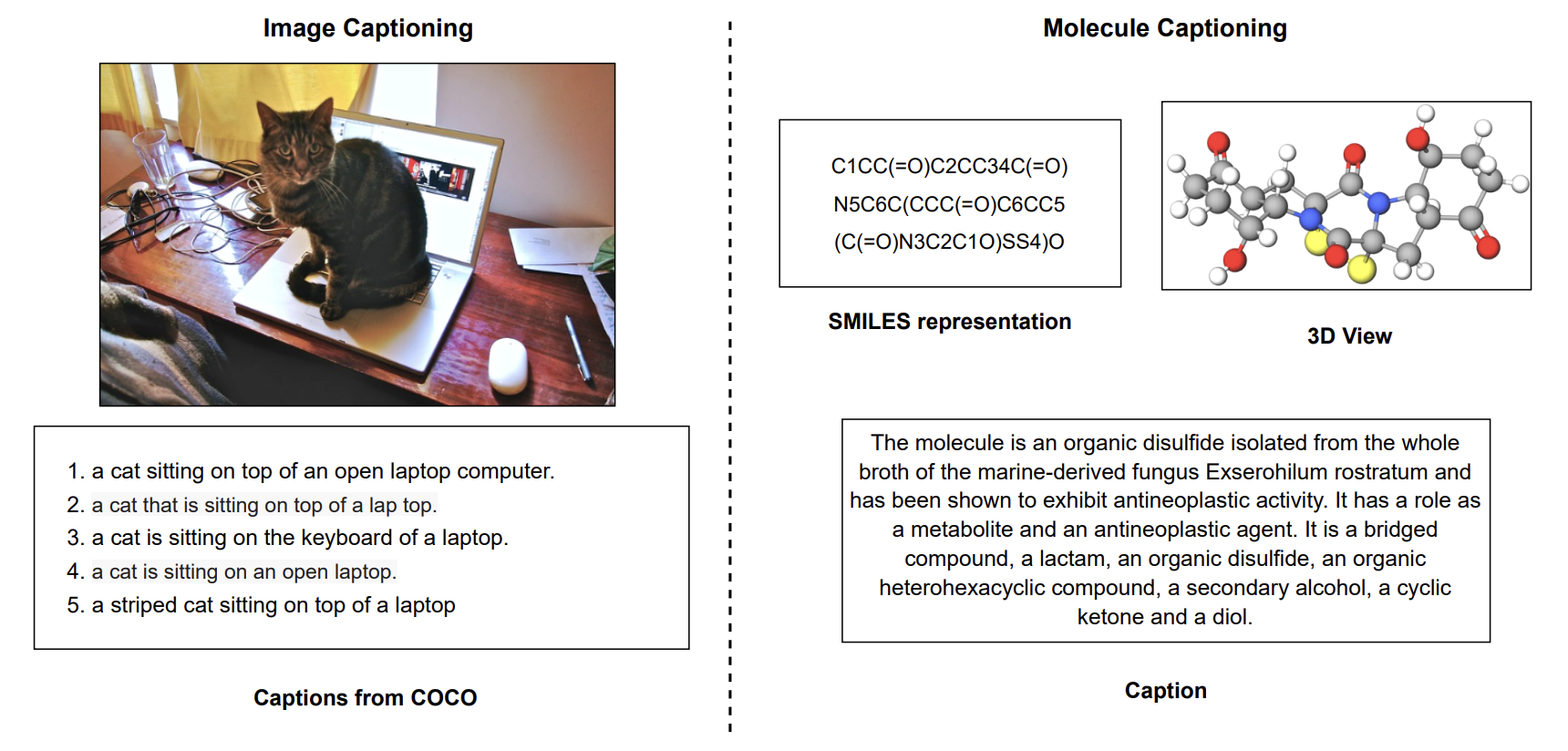
We present MolT5 - a self-supervised learning framework for pretraining models on a vast amount of unlabeled natural language text and molecule strings. MolT5 allows for new, useful, and challenging analogs of traditional vision-language tasks, such as molecule captioning and text-based de novo molecule generation (altogether: translation between molecules and language), which we explore for the first time. Since MolT5 pretrains models on single-modal data, it helps overcome the chemistry domain shortcoming of data scarcity. Furthermore, we consider several metrics, including a new cross-modal embedding-based metric, to evaluate the tasks of molecule captioning and text-based molecule generation. Our results show that MolT5-based models are able to generate outputs, both molecules and captions, which in many cases are high quality.
Software
All code and data for MolT5 can be accessed and downloaded at https://github.com/blender-nlp/MolT5.
Datasets
Evaluation
We provide evaluation code for these new tasks on Github.
Installation
The requirements for the evaluation code conda environment are in environment_eval.yml. An environment can be created using the following commands:
conda env create -n MolTextTranslationEval -f
environment_eval.yml python=3.9
conda activate MolTextTranslationEval
python -m spacy download en_core_web_sm
pip install git+https://github.com/samoturk/mol2vec
Required Downloads for Text2Mol Metric
- test_outputfinal_weights.320.pt should be placed in "evaluation/t2m_output".
It can be downloaded using
curl -L https://uofi.box.com/shared/static/es16alnhzfy1hpagf55fu48k49f8n29x --output test_outputfinal_weights.320.pt
Input format
The input format should be a tab-separated txt file with three columns and the header
'SMILES ground truth output'
'description ground truth output'
Evaluation Commands
| Code | Evaluation |
| Evaluating SMILES to Caption | |
| python text_translation_metrics.py --input_file smiles2caption_example.txt | Evaluate all NLG metrics. |
| python text_text2mol_metric.py --input_file smiles2caption_example.txt | Evaluate Text2Mol metric for caption generation. |
| python text_text2mol_metric.py --use_gt | Evaluate Text2Mol metric for the ground truth. |
| Evaluating Caption to SMILES | |
| python mol_translation_metrics.py --input_file caption2smiles_example.txt | Evaluate BLEU, Exact match, and Levenshtein metrics. |
| python fingerprint_metrics.py --input_file caption2smiles_example.txt | Evaluate fingerprint metrics. |
| ./mol_text2mol_metric.sh caption2smiles_example.txt | Evaluate Text2Mol metric for molecule generation. |
| python mol_text2mol_metric.py --use_gt | Evaluate Text2Mol metric for the ground truth. |
| python fcd_metric.py --input_file caption2smiles_example.txt | Evaluate FCD metric for molecule generation. |
HuggingFace model checkpoints
All of our HuggingFace checkpoints are located here.
Pretrained MolT5-based checkpoints include:
- molt5-small (~77 million parameters)
- molt5-base (~250 million parameters)
- molt5-large (~800 million parameters)
Example usage for molecule captioning (i.e., smiles2caption):
from transformers import T5Tokenizer, T5ForConditionalGeneration
tokenizer = T5Tokenizer.from_pretrained("laituan245/molt5-large-smiles2caption", model_max_length=512)
model = T5ForConditionalGeneration.from_pretrained('laituan245/molt5-large-smiles2caption')
input_text = 'C1=CC2=C(C(=C1)[O-])NC(=CC2=O)C(=O)O'
input_ids = tokenizer(input_text, return_tensors="pt").input_ids
outputs = model.generate(input_ids, num_beams=5, max_length=512)
print(tokenizer.decode(outputs[0], skip_special_tokens=True))
Example usage for molecule generation (i.e., caption2smiles):
from transformers import T5Tokenizer, T5ForConditionalGeneration
tokenizer = T5Tokenizer.from_pretrained("laituan245/molt5-large-caption2smiles", model_max_length=512)
model = T5ForConditionalGeneration.from_pretrained('laituan245/molt5-large-caption2smiles')
input_text = 'The molecule is a monomethoxybenzene that is 2-methoxyphenol substituted by a hydroxymethyl group at position 4. It has a role as a plant metabolite. It is a member of guaiacols and a member of benzyl alcohols.'
input_ids = tokenizer(input_text, return_tensors="pt").input_ids
outputs = model.generate(input_ids, num_beams=5, max_length=512)
print(tokenizer.decode(outputs[0], skip_special_tokens=True))
Acknowledgement
We would like to thank Martin Burke for his helpful discussion. This research is based upon work supported by the Molecule Maker Lab Institute: an AI research institute program supported by NSF under award No. 2019897 and No. 2034562. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein.
References
Carl Edwards, Tuan Lai, Kevin Ros, Garrett Honke, Kyunghyun Cho, Heng Ji. Translation between Molecules and Natural Language. EMNLP 2022.